Fast bindings for LibSVM in scikits.learn
LibSVM is a C++ library that implements several Support Vector Machine algorithms that are commonly used in machine learning. It is a fast library that has no dependencies and most machine learning frameworks bind it in some way or another. LibSVM comes with a Python interface written in swig, but this interface is inherently slow as it does not take into account numpy's array structure. Also, it does not wrap all the library's functionality. Some projects bind it using this bindings and other (such as PyMVPA) make its own wrap, binding some methods directly to numpy's array structure. My approach was to code all algorithms that convert libsvm's data structures (sparse) to numpy arrays (dense) in pure C and wrap them in a very thin Cython layer. Special attention was given to minimize the overhead of converting between libsvm data structures and numpy arrays, as in my opinion this was the main source of bad performance in existing python bindings.
Benchmarks
As a first benchmark, I supposed a situation in which the dimension of the subspace is small and there are lots of points to classify. This is typically the case when your data is points in plane or in space and you want to draw the decision function by classifying every point in the grid. In this case, the bottleneck is not the classification algorithm, but the conversion of data from a dense representation used by python and numpy and a sparse representation used by libsvm. Not surprisingly, we get huge performance gains if we speed up the conversion dense/sparse.

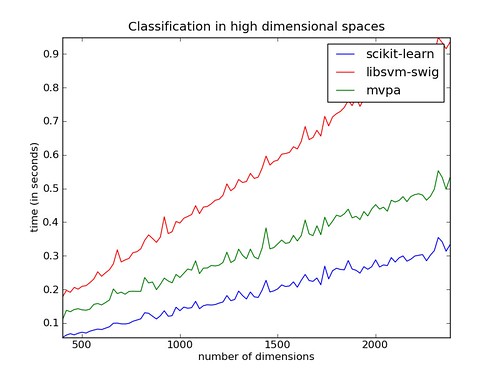
Curse of dimensionality
In the case of a huge number of dimensions, the speedup is not so spectacular, but we also get a performance boost by making training somewhat faster.
Bidirectional mapping
A feature that was needed and that I haven't found on other implementations is that you can tweak parameters in the SVM class and the classifier will reflect those changes (i.e. parameters are actually copied back and forth, not just passed as an opaque pointer). Suppose you train an instance of the classifier and are interested in the coefficients that multiply the support vectors in the decision function. In scikits.learn, you can access this array under field .coef_: `` >>> import numpy as np >>> from scikits.learn import svm >>> clf = svm.SVM() >>> clf.fit([[1,2], [3,4]], [-1, 1]) >>> clf.coef clf.coef0 clf.coef_ >>> clf.coef_ array([[ 1., -1.]])`` Now, changing the value of these coefficients effectively changes the decision function: `` >>> clf.predict([[1,2]]) array([ -1.]) >>> clf.coef_ = np.array([[0.0, -1.0]]) >>> clf.predict([[1,2]]) array([ 1.])``
Code
All code can be found in the scikit (you'll have to get the svn version), in file scikits/learn/svm.py and scikits/learn/src/. All plots are generated from this script
Notes
In the benchmarks, a Linear Kernel was used, as it is the most common. Other more computationally intensive kernels would probably narrow the difference.
Bugs
This code should be treated as alpha quality and has not being extensively tested. Please report any bugs that you encounter to the tracker